RNA Modifications & Structure Profiling
Structure profiling and natural modification detection with UltraMarathonRT


Structure profiling and natural modification detection with UltraMarathonRT

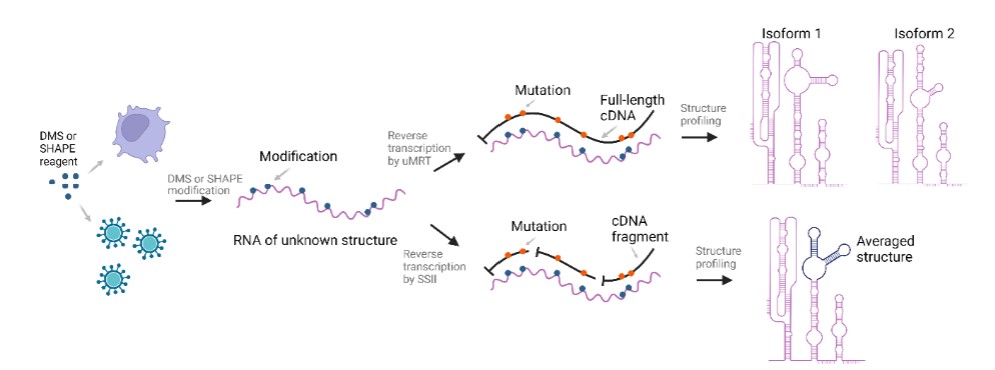
UltraMarathonRT (uMRT) is highly sensitive to the modifications deposited to RNA by DMS and SHAPE reagents (such as NAI and 2A3), and thus it has been widely adopted to profile RNA secondary structures using DMS-MaP and SHAPE-MaP. Due to its ultra-high processivity and ability to unwind RNA structures, uMRT can copy any RNA template in a single pass, making it the ideal reverse transcriptase (RT) enzyme for this application. Furthermore, it is the only ultra-processive RT that offers single-molecule resolution. uMRT can enhance the accuracy, coverage, and depth of findings for your RNA structural studies.
1. Ultra-Processivity providing single-molecule resolution
UltraMarathonRT is the ultimate solution for efficient, full-length cDNA synthesis from
long RNA templates. It has been engineered to bind tightly to RNA templates,
ensuring minimal dissociation and exceptional end-to-end processivity, making it
ideal for RNA structure profiling. Combining the full-length cDNA synthesized by
uMRT with nanopore long-read sequencing enables accurate, in-depth structural
analysis that provides visibility into distinct structural isoforms which cannot be
discerned using traditional, distributive RTs (ref: Nature Methods (2023) 20,
849–859 and Nature Protocols
(2024) 19, 1835–1865).
2 Low Error Rates
UltraMarathonRT offers superior accuracy with lower background error rates
compared to traditional reverse transcriptases. By minimizing false-positive
mutations, uMRT provides reliable detection of chemical modifications and precise
analysis of RNA structural features, ensuring more accurate results in your research.
3. Decoding Modifications on all four RNA bases in DMS-MaP
UltraMarathonRT generates unique mutational signatures on DMS modified uracil
(U) and guanine (G), making it the only RT enzyme that can accurately detect DMS-
induced modifications on all four RNA bases—adenine, cytosine, uracil, and guanine
(ref:
Nucleic Acids Research (2023) 51,
8744–8757). This expands RNA structural
profiling, offering comprehensive nucleotide-level insights, even for previously
challenging bases like guanine.
4. Single-Molecule Resolution
UltraMarathonRT enables more sensitive and high-resolution single-molecule
analysis. This is crucial for detecting correlated modification events, which are used
to infer secondary and tertiary structures, as well as dynamic structural states in
RNA molecules.
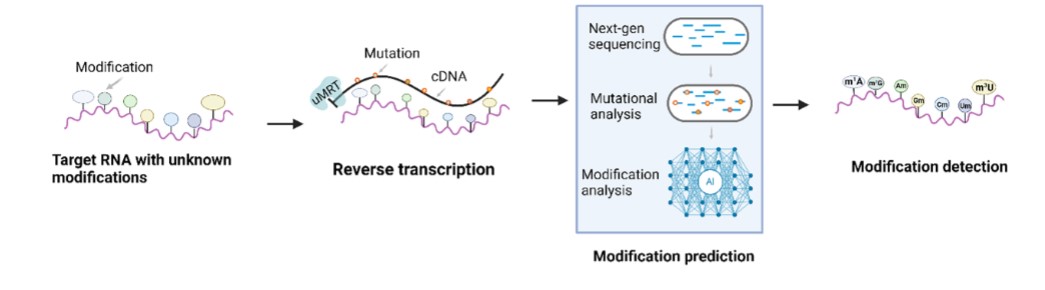
5. Natural Modification Detection
UltraMarathonRT is also sensitive to other RNA modifications, making it the first
epitranscriptomic tool that detects post-transcriptional modifications (ref: Journal of
Molecular Biology (2023), 435, 168299).